Detailed knowledge concerning your genetic makeup only change an estimated disease prediction risk for three common diseases by a few percentage points which is insufficient to make a treatment plans.

The study appears online May 24, 2012 and will appear in the June 8, 2012 print issue of The American Journal of Human Genetics.
Scientists have long hoped that using genetic information gleaned from the Human Genome Project and other genetic research could improve disease risk prediction enough to help aid in prevention and treatment. Others have been skeptical that such "personalized medicine" will be of clinical benefit. Still others have argued that there will be benefits in the future, but that current risk prediction algorithms underperform because they don't allow for potential synergistic effects—the interplay of multiple genetic risk markers and environmental factors—instead focusing only on individual genetic markers.
Aschard and his co-authors, including senior author Peter Kraft, HSPH associate professor of epidemiology, examined whether disease risk prediction would improve for breast cancer, type 2 diabetes, and rheumatoid arthritis if they included the effect of synergy in their statistical models. But they found no significant effect by doing so. "Statistical models of synergy among genetic markers are not 'game changers' in terms of risk prediction in the general population," said Aschard.
The researchers conducted a simulation study by generating a broad range of possible statistical interactions among common environmental exposures and common genetic risk markers related to each of the three diseases. Then they estimated whether such interactions would significantly boost disease prediction risk when compared with models that didn't include these interactions since, to date, using individual genetic markers in such predictions has provided only modest improvements.
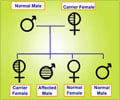
For breast cancer, the researchers considered 15 common genetic variations associated with disease risk and environmental factors such as age of first menstruation, age at first birth, and number of close relatives who developed breast cancer. For type 2 diabetes, they looked at 31 genetic variations along with factors such as obesity, smoking status, physical activity, and family history of the disease. For rheumatoid arthritis, they also included 31 genetic variations, as well as two environmental factors: smoking and breastfeeding.
Advertisement
"Overall, our findings suggest that the potential complexity of genetic and environmental factors related to disease will have to be understood on a much larger scale than initially expected to be useful for risk prediction. The road to efficient genetic risk prediction, if it exists, is likely to be long," said Aschard.
Advertisement
Still, Aschard and his co-authors recommend further study of gene-gene and gene-environmental interactions because it can provide important clues, if not about disease risk, at least about disease causes—which could in turn lead to improved treatment and prevention strategies.
Marilyn Cornelis, research associate in the Department of Nutrition at HSPH, was also a co-author on the study.
Source-Eurekalert