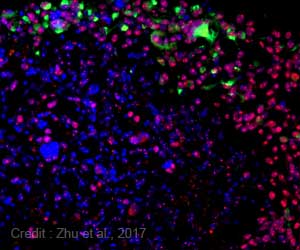
‘University of Texas (UT) Southwestern Medical Center researchers used three endocytic pathways as markers to sort out 29 non-small cell lung cancer (NSCLC) lines into two distinct clusters characterized by their endocytic dysfunction.’
Tweet it Now
Endocytosis is not normal in cancer cells but how dysregulated the process is in cancer cells has just been revealed by Sarah Elkin and colleagues in the University of Texas (UT) Southwestern Medical Center lab of Sandra Schmid. The researchers used three endocytic pathways as markers to sort out 29 non-small cell lung cancer (NSCLC) lines into two distinct clusters characterized by their endocytic dysfunction. Elkin will present Tuesday, December 15, at ASCB 2015 in San Diego on the work published1 last month in Cancer Research. For cancer to progress, cancer cells modify normal signaling, movement, and growth processes. These cell activities are regulated by proteins normally found at the cell's membrane. A cell can change the amount of these cell surface proteins by internalizing small portions of the membrane through endocytosis. The UT Southwestern researchers monitored endocytosis in NSCLCs and found significant heterogeneity among the various cell lines, underlining the astounding variety and personal nature of cancer types.
But by hierarchical clustering of observed alterations in endocytosis, the researchers identified two distinct cancer phenotype clusters, one marked by mutations in the oncogene KRAS in mesenchymal cells and the other by changes in epithelial cells.
Their results, say the UT Southwestern researchers, show that changes in endocytosis reflected by the expression of important molecules on the cell surface can be used to distinguish cancer subtypes. But they also warn that their work shows the danger of drawing general conclusions about cancer-relevant phenotypes from a single cancer cell line. Cancer can be chillingly personal, right down to your cell surface.
Source-Eurekalert